16 Chapter 16: Prokaryotic Gene Expression
Lisa Limeri; Joshua Reid; and rocksher
Learning Objectives
By the end of this section, you will be able to do the following:
-
Describe the idea of “differential gene expression” by identifying the different possible regulatory regions of a gene and distinguishing between the roles of each region for regulating gene expression, including transcriptional, post-transcriptional, and translational regulation.
-
Predict the most likely effect of mutations in different regulatory regions on gene expression (transcription, post-transcription, and translation).

Introduction
For a cell to function properly, necessary proteins must be synthesized at the proper time and place. All cells control or regulate the synthesis of proteins from information encoded in their DNA. The process of turning on a gene to produce RNA and protein is called gene expression. Whether in a simple unicellular organism or a complex multi-cellular organism, the expression of individual genes, or groups of genes, is tightly regulated by the cell. For this to occur, there must be internal chemical mechanisms that control when a gene is expressed to make RNA and protein, how much of the protein is made, and when it is time to stop making that protein because it is no longer needed.
The regulation of gene expression conserves energy and space. It would require a significant amount of energy for an organism to express every gene at all times, so it is more energy efficient to turn on the genes only when they are required. In addition, only expressing a subset of genes in each cell saves space because DNA must be unwound from its tightly coiled structure to transcribe and translate the DNA. Cells would have to be enormous if every gene were expressed in every cell all the time. The control of gene expression is extremely complex. Malfunctions in this process are detrimental to the cell and can lead to the development of many diseases, including cancer.
Regulation of Gene Expression in Prokaryotes
To understand how gene expression is regulated, we must first understand how a gene codes for a functional protein in a cell. The process occurs in both prokaryotic and eukaryotic cells, just in slightly different manners.
Prokaryotic organisms are single-celled organisms that lack a cell nucleus, and their DNA therefore floats freely in the cell cytoplasm. To synthesize a protein, the processes of transcription and translation occur almost simultaneously. When the resulting protein is no longer needed, transcription stops. As a result, the primary method to control what type of protein and how much of each protein is expressed in a prokaryotic cell is the regulation of DNA transcription (Fig 18.1). All of the subsequent steps occur automatically. When more protein is required, more transcription occurs. Therefore, in prokaryotic cells, the control of gene expression is mostly at the transcriptional level.
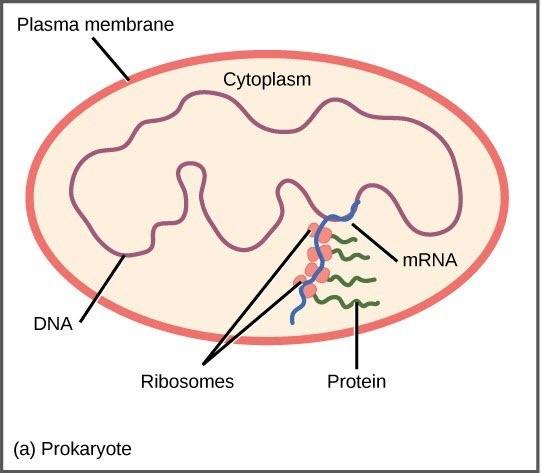
The DNA of prokaryotes is organized into a circular chromosome, supercoiled within the nucleoid region of the cell cytoplasm. Genes for proteins that are needed for a specific function, or that are involved in the same biochemical pathway, are clustered together in blocks called operons. For example, all of the genes needed to use lactose as an energy source are coded next to each other in the lactose (or lac) operon, and transcribed into a single mRNA.
In prokaryotic cells, there are three types of regulatory molecules that can affect the expression of operons: repressors, activators, and inducers. Repressors and activators are proteins produced in the cell. Both repressors and activators regulate gene expression by binding to specific DNA sites adjacent to the genes they control. In general, activators bind to the promoter site, while repressors bind to operator regions. Repressors prevent transcription of a gene in response to an external stimulus, whereas activators increase the transcription of a gene in response to an external stimulus. Inducers are small molecules that may be produced by the cell or that are in the cell’s environment. Inducers either activate or repress transcription depending on the needs of the cell and the availability of substrate.
Reading Question #1
What is the process by which a gene is turned on to produce RNA and protein called?
A. Protein synthesis
B. Transcription
C. Translation
D. Gene expression
Reading Question #2
In prokaryotic cells, where does DNA transcription and translation occur?
A. Nucleus
B. Mitochondria
C. Cytoplasm
D. Endoplasmic reticulum
The lac Operon: An Inducible Operon
One type of gene regulation in prokaryotic cells occurs through inducible operons, which have proteins that bind to activate transcription depending on the local environment and the needs of the cell. The lac operon is a typical inducible operon found in the bacterium Escherichia coli. When glucose concentrations are low, E. coli is able to use other sugars, like lactose, as energy sources. The lac operon encodes the genes necessary to acquire and process the lactose from the local environment. The Z gene of the lac operon encodes beta-galactosidase, which breaks lactose down to glucose and galactose.
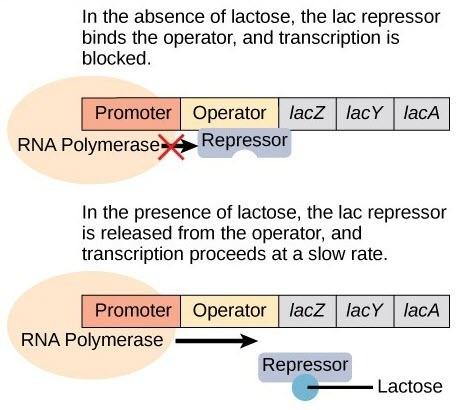
Expression of the lac operon is turned off under normal conditions (high glucose) due to the binding of the lac repressor protein (Figure 18.2). Only when glucose is absent and lactose is present will the lac operon be transcribed (Figure 18.2). When lactose is present, its metabolite, allolactose, binds to the lac repressor and changes its shape so that it cannot bind to the lac operator to prevent transcription. RNA polymerase is now free to initiate transcription of the lac operon in response the the presence of lactose.
The lac repressor stands between the cell and the enzymes needed for lactose processing. The expression of the operon continues until sufficient amounts of protein are produced. The allolactose is a victim of its own success and is targeted by enzymes and rendered inactive. The lac operon returns to its default state. This combination of conditions makes sense for the cell, because it would be energetically wasteful to synthesize the enzymes to process lactose if glucose was plentiful or lactose was not available. It should be mentioned that the lac operon is transcribed at a very low rate even when glucose is present and lactose absent.
Reading Question #3
When is the lac operon transcribed in E. coli?
A. When glucose is absent and lactose is present.
B. When glucose is present and lactose is absent.
C. When glucose and lactose are both present.
D. When glucose and lactose are both absent.
Reading Question #4
What is the function of the lac operon in E. coli?
A. To break down glucose into lactose and galactose.
B. To bind to the lac repressor protein and prevent transcription.
C. To regulate the expression of genes involved in lactose metabolism.
D. To synthesize allolactose for regulating glucose concentration.
The trp Operon: A Repressible Operon
Bacteria such as E. coli need amino acids to survive, and are able to synthesize many of them. Tryptophan is one such amino acid that E. coli can either ingest from the environment or synthesize using enzymes that are encoded by five genes. These five genes are next to each other in what is called the tryptophan (trp) operon (Figure 18.3). The genes are transcribed into a single mRNA, which is then translated to produce all five enzymes. If tryptophan is present in the environment, then E. coli does not need to synthesize it and the trp operon is switched off by default. However, when tryptophan availability is low, the switch controlling the operon is turned on, the mRNA is transcribed, the enzyme proteins are translated, and tryptophan is synthesized.
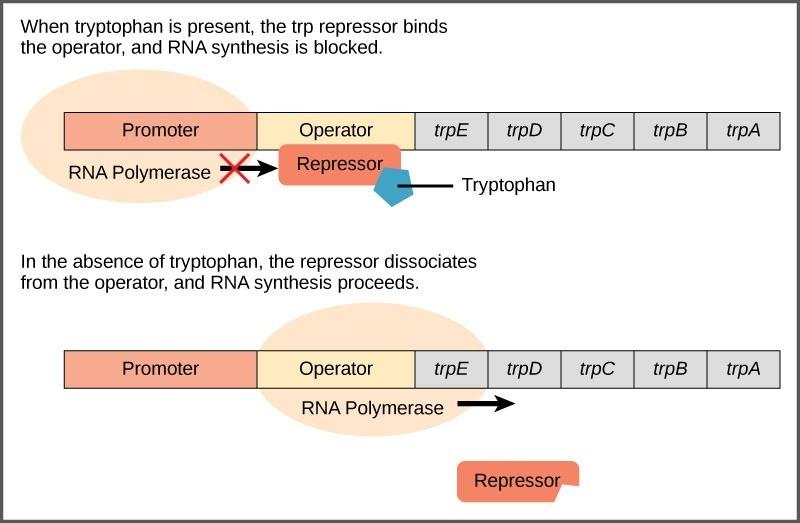
The trp operon includes three important regions: the coding region, the trp operator and the trp promoter. The coding region includes the genes for the five tryptophan biosynthesis enzymes. Just before the coding region is the transcriptional start site. The promoter sequence, to which RNA polymerase binds to initiate transcription, is before or “upstream” of the transcriptional start site. Between the promoter and the transcriptional start site is the operator region.
The trp operator contains the DNA code to which the trp repressor protein can bind. However, the repressor alone cannot bind to the operator. When tryptophan is present in the cell, two tryptophan molecules bind to the trp repressor, which changes the shape of the repressor protein to a form that can bind to the trp operator. Binding of the tryptophan–repressor complex at the operator physically prevents the RNA polymerase from binding to the promoter and transcribing the downstream genes.
When tryptophan is not present in the cell, the repressor by itself does not bind to the operator, the polymerase can transcribe the enzyme genes, and tryptophan is synthesized. Because the repressor protein actively binds to the operator to keep the genes turned off, the trp operon is said to be negatively regulated and the proteins that bind to the operator to silence trp expression are negative regulators.
Reading Question #5
What is the function of the trp operon in E. coli?
A. To break down tryptophan into its component amino acids.
B. To regulate the expression of genes involved in tryptophan biosynthesis.
C. To prevent the binding of RNA polymerase to the promoter sequence.
D. To synthesize tryptophan from available amino acids.

References and Acknowledgements
Adapted from Clark, M.A., Douglas, M., and Choi, J. (2018). Biology 2e. OpenStax. Retrieved from https://openstax.org/books/biology-2e/pages/1-introduction

