5 Chapter 5: DNA Replication
Lisa Limeri; Joshua Reid; rocksher; and Shifath Bin Syed
Learning Objectives
By the end of this section, you will be able to do the following:
- Use a drawing that you create to explain the statement: “A newly synthesized DNA strand is half old and half new.”
- Given a diagram of a DNA molecule during replication, label the following: the origin of replication, directions of replication, replication fork, the leading strand, and lagging strands and their polarities, and the replisome.
- Describe the function of major components of the replisome: helicase, topoisomerase, DNA polymerase, DNA ligase, and primase.
Introduction
The three letters “DNA” have now become synonymous with crime-solving and genetic testing. DNA can be retrieved from hair, blood, or saliva. Each person’s DNA is unique, and it is possible to detect individual differences within a species based on these unique features.
DNA analysis has many practical applications beyond forensics. In humans, DNA testing is applied to numerous uses: determining paternity, tracing genealogy, identifying pathogens, archaeological research, tracing disease outbreaks, and studying human migration patterns. In the medical field, DNA is used in diagnostics, new vaccine development, and cancer therapy. It is now possible to determine predisposition to diseases by looking at genes.
Each human cell has 23 pairs of chromosomes: one set is inherited from the female parent, and the other is inherited from the male parent. A mitochondrial genome, inherited exclusively from the female parent, can be involved in inherited genetic disorders. On each chromosome, thousands of genes are responsible for determining the genotype and phenotype of the individual. A gene is defined as a sequence of DNA that codes for a functional product. The human haploid genome contains 3 billion base pairs and has between 20,000 and 25,000 functional genes.
Basics of DNA Replication
The elucidation of the structure of the double helix provided a hint as to how DNA divides and makes copies of itself. In their 1953 paper, Watson and Crick penned an incredible understatement: “It has not escaped our notice that the specific pairing we have postulated immediately suggests a possible copying mechanism for the genetic material.” With specific base pairs, the sequence of one DNA strand can be predicted from its complement. The double-helix model suggests that the two strands of the double helix separate during replication, and each strand serves as a template from which the new complementary strand is copied. What was not clear was how the replication took place. There were three models suggested (Fig 5.1): conservative, semi-conservative, and dispersive.

In conservative replication, the parental DNA remains together, and the newly formed daughter strands are together. The semi-conservative method suggests that each of the two parental DNA strands acts as a template for new DNA to be synthesized; after replication, each double-stranded DNA includes one parental or “old” strand and one “new” strand. In the dispersive model, both copies of DNA have double-stranded segments of parental DNA and newly synthesized DNA interspersed.
Meselson and Stahl were interested in understanding how DNA replicates. They grew E. coli for several generations in a medium containing a “heavy” isotope of nitrogen (15N), which gets incorporated into nitrogenous bases, and eventually into the DNA. The E. coli culture was then placed into medium containing 14N and allowed to grow for several generations. After each of the first few generations, the cells were harvested and the DNA was isolated, then centrifuged at high speeds in an ultracentrifuge. During the centrifugation, the DNA was loaded into a gradient and spun at high speeds. Under these circumstances, the DNA will form a band according to its buoyant density: the density within the gradient at which it floats. DNA grown in 15N will form a band at a higher density position (i.e., farther down the centrifuge tube) than that grown in 14N. Meselson and Stahl noted that after one generation of growth in 14N after they had been shifted from 15N, the single band observed was intermediate in position in between DNA of cells grown exclusively in 15N and 14N. This suggested either a semi-conservative or dispersive mode of replication. The DNA harvested from cells grown for two generations in 14N formed two bands: one DNA band was at the intermediate position between 15N and 14N, and the other corresponded to the band of 14N DNA. These results could only be explained if DNA replicates in a semi-conservative manner. And for this reason, therefore, the other two models were ruled out.
Reading Check #1
One strand of the original DNA pair is incorporated into each of the new strands during replication. This type of replication is described as:
A. Conservative
B. Liberal
C. Reactionary
D. Semi-conservative
DNA Replication in Prokaryotes
DNA replication has been well studied in prokaryotes, primarily because of the small genome size and the large variety of available mutants. E. coli has 4.6 million base pairs in a single circular chromosome. All of it gets replicated in approximately 42 minutes, starting from a single site along the chromosome and proceeding around the circle in both directions. This means that approximately 1,000 nucleotides are added per second. Thus, the process is quite rapid and occurs without many mistakes.
DNA replication employs a large number of structural proteins and enzymes, each of which plays a critical role during the process. One of the key players is the enzyme DNA polymerase, also known as DNA pol, which adds nucleotides one by one to the growing DNA chain that is complementary to the template strand. Adding nucleotides requires energy obtained from the nucleoside triphosphates ATP, GTP, TTP, and CTP. Like ATP, the other NTPs (nucleoside triphosphates) are high-energy molecules that can serve as the source of DNA nucleotides and energy to drive polymerization. When the bond between the terminal phosphate is cleaved off the NTP, the energy released powers the formation of the phosphodiester bond between the incoming nucleotide and the growing chain. In prokaryotes, three main types of polymerases are known: DNA pol I, DNA pol II, and DNA pol III. It is now known that DNA pol III is the enzyme required for DNA synthesis (Fig 5.1); DNA pol I is an important accessory enzyme in DNA replication and, along with DNA pol II, is primarily required for repair.
How does the replication machinery know where to begin? It turns out that there are specific nucleotide sequences called origins of replication where replication begins. In E. coli, which has a single origin of replication on its one chromosome (as do most prokaryotes), this origin of replication is approximately 245 base pairs long and is rich in AT sequences. The origin of replication is recognized by certain proteins that bind to this site. An enzyme called helicase unwinds the DNA by breaking the hydrogen bonds between the nitrogenous base pairs. ATP hydrolysis is required for this process. As the DNA opens up, Y-shaped structures called replication forks are formed. Two replication forks are formed at the origin of replication, which is extended bi-directionally as replication proceeds. Single-strand binding proteins coat the single strands of DNA near the replication fork to prevent the single-stranded DNA from rewinding into a double helix.
DNA polymerase has two important restrictions: it is able to add nucleotides only in the 5′ to 3′ direction (a new DNA strand can only be extended in this direction). It also requires a free 3′-OH group to which it can add nucleotides by forming a phosphodiester bond between the 3′-OH end and the 5′ phosphate of the next nucleotide. This essentially means that it cannot add nucleotides if a free 3′-OH group is not available. Then how does it add the first nucleotide? The problem is solved with the help of a primer that provides a free 3’OH end. Another enzyme, RNA primase, synthesizes an RNA segment that is about five to ten nucleotides long and complementary to the template DNA. Because this sequence primes the DNA synthesis, it is appropriately called the primer. DNA polymerase can now extend this RNA primer, adding nucleotides one by one that is complementary to the template strand (Fig 5.1).
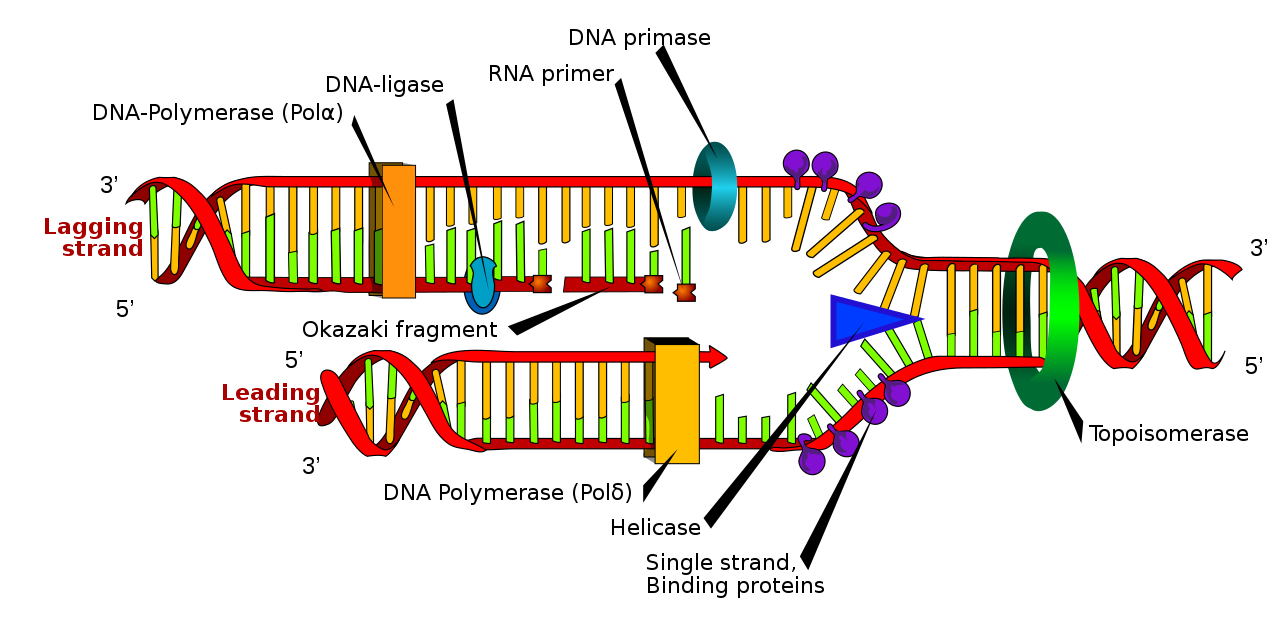
The replication fork moves at a rate of 1,000 nucleotides per second. Topoisomerase prevents the overwinding of the DNA double helix ahead of the replication fork as the DNA is opening up; it does so by causing temporary nicks in the DNA helix and then resealing it. Because DNA polymerase can only extend in the 5′ to 3′ direction, and because the DNA double helix is antiparallel, there is a slight problem at the replication fork. The two template DNA strands have opposing orientations: one strand is in the 5′ to 3′ direction, and the other is oriented in the 3′ to 5′ direction. Only one new DNA strand, the one that is complementary to the 3′ to 5′ parental DNA strand, can be synthesized continuously towards the replication fork (Fig 5.2). This continuously synthesized strand is known as the leading strand. The other strand, complementary to the 5′ to 3′ parental DNA, is extended away from the replication fork in small fragments known as Okazaki fragments, each requiring a primer to start the synthesis. New primer segments are laid down in the direction of the replication fork, each pointing away from it. Okazaki fragments are named after the Japanese scientist who first discovered them. The strand with the Okazaki fragments is known as the lagging strand.
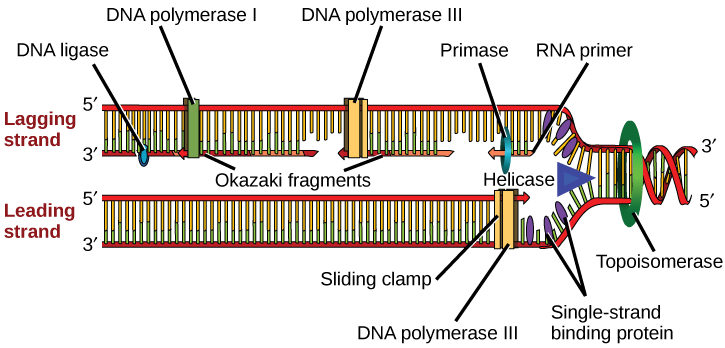
The leading strand can be extended with a single primer, whereas the lagging strand needs a new primer for each short Okazaki fragment. A protein called the sliding clamp holds the DNA polymerase in place as it adds nucleotides. The sliding clamp is a ring-shaped protein that binds to the DNA and holds the polymerase in place. As synthesis proceeds, the RNA primers are replaced by DNA Pol I. The primers are removed by the exonuclease activity of DNA pol I, which uses DNA behind the RNA as its primer and fills in the gaps left by the removal of the RNA nucleotides with the addition of DNA nucleotides. The nicks that remain between the newly synthesized DNA (that replaced the RNA primer) and the previously synthesized DNA are sealed by the enzyme DNA ligase, which catalyzes the formation of phosphodiester linkages between the 3′-OH end of one nucleotide and the 5′ phosphate end of the other fragment.
The process of DNA replication can be summarized as follows (review the full process of DNA replication here as well):
- DNA unwinds at the origin of replication.
- Helicase opens up the DNA-forming replication forks; these are extended bidirectionally.
- Single-strand binding proteins coat the DNA around the replication fork to prevent rewinding of the DNA.
- Topoisomerase binds to the region ahead of the replication fork to prevent supercoiling.
- Primase synthesizes RNA primers complementary to the DNA strand.
- DNA polymerase III starts adding nucleotides to the 3′-OH end of the primer.
- Elongation of both the lagging and the leading strands continues.
- RNA primers are removed by exonuclease activity.
- Gaps are filled by DNA pol I by adding dNTPs.
- The nick between the two DNA fragments is sealed by DNA ligase, which helps in the formation of phosphodiester bonds.
Table 5.1 summarizes the enzymes involved in prokaryotic DNA replication and the functions of each.
Table 5.1 Prokaryotic DNA Replication: Enzymes and Their Function
|
Enzyme/protein |
Specific Function |
|
DNA pol I |
Removes RNA primer and replaces it with newly synthesized DNA |
|
DNA pol III |
Main enzyme that adds nucleotides in the 5′-3′ direction |
|
Helicase |
Opens the DNA helix by breaking hydrogen bonds between the nitrogenous bases |
|
Ligase |
Seals the nick between the Okazaki fragments to create one continuous DNA strand |
|
Primase |
Synthesizes RNA primers needed to start replication |
|
Sliding Clamp |
Helps to hold the DNA polymerase in place when nucleotides are being added |
|
Topoisomerase |
Helps relieve the strain on DNA when unwinding by causing breaks, and then resealing the DNA |
|
Single-strand binding proteins (SSB) |
Binds to single-stranded DNA to prevent DNA from rewinding back. |
Reading Check #2
Which enzyme is responsible for adding nucleotides one by one to the growing DNA chain during replication?
A. Helicase
B. DNA polymerase
C. Primase
D. RNA polymerase
Reading Check #3
What is the function of the RNA primer in DNA replication?
A. To unwind the DNA strands.
B. To stabilize the single-stranded DNA.
C. To provide a free 3′-OH end for DNA polymerase.
D. To break the hydrogen bonds between the nitrogenous base pairs.
DNA Replication in Eukaryotes
Eukaryotic genomes are much more complex and larger in size than prokaryotic genomes. Eukaryotes also have several different linear chromosomes. The human genome has 3 billion base pairs per haploid set of chromosomes, and 6 billion base pairs are replicated during the S phase of the cell cycle. Each eukaryotic chromosome has multiple origins of replication; humans can have up to 100,000 origins of replication across the genome. The replication rate is approximately 100 nucleotides per second, which is much slower than prokaryotic replication. In yeast, which is a eukaryote, special sequences known as autonomously replicating sequences (ARS) are found on the chromosomes. These are equivalent to the origin of replication in E. coli.
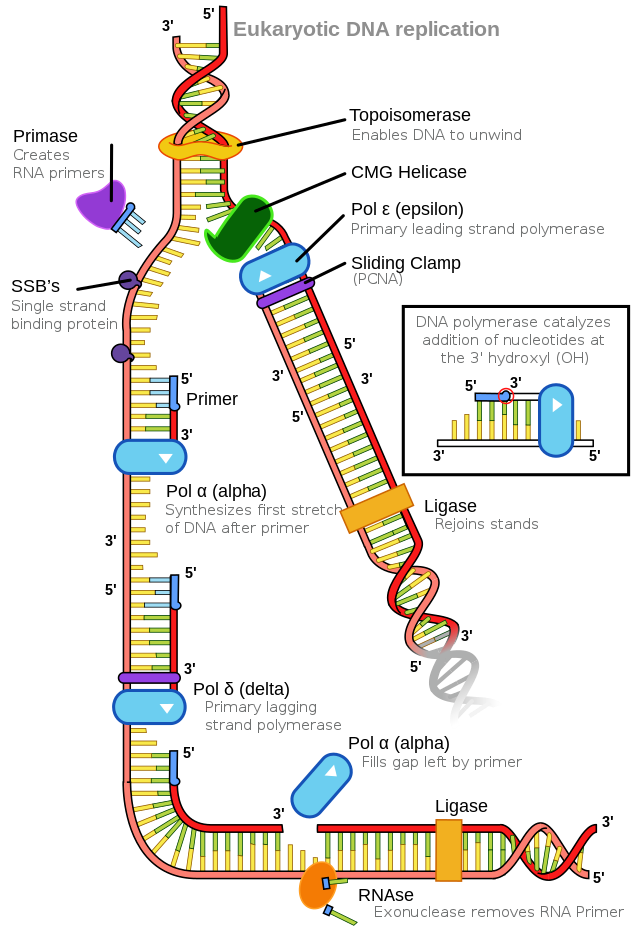
The number of DNA polymerases in eukaryotes is much higher than in prokaryotes: 14 are known, of which five are known to have major roles during replication and have been well studied. They are known as pol α, pol β, pol γ, pol δ, and pol ε.
The essential steps of replication are the same as in prokaryotes. Before replication can start, the DNA has to be made available as a template. Eukaryotic DNA is bound to basic proteins known as histones to form nucleosome structures. Histones must be removed and then replaced during the replication process, which helps to account for the slower replication rate in eukaryotes. The chromatin may undergo some chemical modifications so that the DNA may be able to slide off the proteins or be accessible to the enzymes of the DNA replication machinery. At the origin of replication, a pre-replication complex is made with other initiator proteins. Helicase and other proteins are recruited to start replication (Table 5.2).
Table 5.2 Difference between Prokaryotic and Eukaryotic Replication
|
Property |
Prokaryotes |
Eukaryotes |
|
Origin of replication |
Single |
Multiple |
|
Rate of replication |
1000 nucleotides/s |
50 to 100 nucleotides/s |
|
DNA polymerase types |
5 |
14 |
|
Telomerase |
Not present |
Present |
|
RNA primer removal |
DNA pol I |
RNase H |
|
Strand elongation |
DNA pol III |
Pol α, pol δ, pol ε |
|
Sliding clamp |
Sliding clamp |
PCNA |
A helicase using the energy from ATP hydrolysis opens up the DNA helix. Replication forks are formed at each replication origin as the DNA unwinds. The opening of the double helix causes over-winding, or supercoiling, in the DNA ahead of the replication fork. These are resolved by the action of topoisomerases. RNA primers are formed by the enzyme primase, and using the primer, DNA pol can start synthesis. Three major DNA polymerases are then involved: α, δ, and ε. DNA pol α adds a short (20 to 30 nucleotides) DNA fragment to the RNA primer on both strands and then hands it off to a second polymerase. While the leading strand is continuously synthesized by the enzyme pol δ, the lagging strand is synthesized by pol ε. A sliding clamp protein known as PCNA (proliferating cell nuclear antigen) holds the DNA pol in place so that it does not slide off the DNA. As pol δ runs into the primer RNA on the lagging strand, it displaces it from the DNA template. The displaced primer RNA is then removed by RNase H and replaced with DNA nucleotides. The Okazaki fragments in the lagging strand are joined after replacing the RNA primers with DNA. The remaining gaps are sealed by DNA ligase, which forms the phosphodiester bond.
Reading Check #4
What is the function of the sliding clamp protein PCNA during DNA replication in eukaryotes?
A. It resolves supercoiling in DNA
B. It forms the phosphodiester bond between DNA fragments
C. It holds DNA polymerase in place
D. It opens up the DNA helix using ATP hydrolysis
Reading Check #5
Which enzyme synthesizes an RNA primer that is complementary to the template DNA during DNA replication?
A. Helicase
B. DNA polymerase
C. Primase
D. RNA Polymerase
References and Acknowledgements
Jaskelioff, M., Muller, F., Paik, J.H., Thomas, E., Jiang, S. … DePinho, R.A. (2011). Telomerase reactivation reverses tissue degeneration in aged telomerase-deficient mice. Nature 469: 102–106. https://doi.org/10.1038/nature09603
Adapted from Clark, M.A., Douglas, M., and Choi, J. (2018). Biology 2e. OpenStax. Retrieved from https://openstax.org/books/biology-2e/pages/1-introduction

