21 Chapter 21: Protein Structure and Folding
Lisa Limeri
Learning Objectives
By the end of this section, students will be able to:
- Label the four components of an amino acid and explain the role of each in terms of how the molecule functions in a protein.
-
Describe each of the four levels of protein structure and explain how each influences the protein’s final size, shape, and chemical properties.
-
Compare which bonds are responsible for producing a protein’s 1) primary structure, 2) secondary structure (alpha-helices and beta-pleated sheets), and 3) tertiary structure.
-
Predict whether the R-group on an amino acid that you haven’t seen before will 1) interact with water, and 2) act as an acid (proton donor) or base (proton acceptor).
Introduction
Proteins are one of the most abundant organic molecules in living systems and have the most diverse range of functions of all macromolecules. Proteins may be structural, regulatory, contractile, or protective. They may serve in transport, storage, or membranes; or they may be toxins or enzymes. Each cell in a living system may contain thousands of proteins, each with a unique function. Their structures, like their functions, vary greatly.
They are all, however, amino acid polymers arranged in a linear sequence.
Proteins fold into complex three-dimensional structures in order to function (Fig 21.1). Our understanding of protein structure and function requires foundational knowledge of chemical bonds. We previously learned about covalent bonds (polar and non-polar), as well as hydrogen bonds when we discussed DNA structure. Please make sure to review the section on chemical bonding if needed. In this section, we will discuss a third type of bond, the ionic bond.

![]()
Ions and Ionic Bonds
Some atoms are more stable when they gain or lose an electron (or possibly two) and form ions. Because the number of electrons does not equal the number of protons, each ion has a net charge. Scientists refer to this movement of electrons from one element to another as electron transfer.
As Figure 21.2 illustrates, sodium (Na) only has one electron in its outer electron shell. It takes less energy for sodium to donate that one electron than it does to accept seven more electrons to fill the outer shell. If sodium loses an electron, which is negatively charged, it then has an overall charge of +1. Similarly, Chlorine (Cl) has seven electrons in its outer shell, and it is more energy-efficient for chlorine to gain one electron than to lose seven. Therefore, it tends to gain a negatively charged electron and gain a net negative (–1) charge. In this example, sodium will donate its one electron to empty its shell, and chlorine will accept that electron to fill its shell. Note that these transactions can normally only take place simultaneously: in order for a sodium atom to lose an electron, it must be in the presence of a suitable recipient like a chlorine atom.
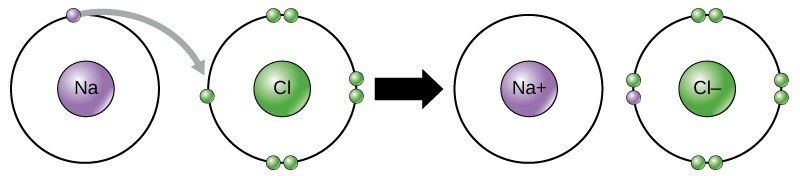
Ionic bonds, a type of electrostatic interaction, form between ions with opposite charges. For instance, positively charged sodium ions and negatively charged chloride ions bond together to make crystals of sodium chloride, or table salt, creating a crystalline molecule with zero net charge.
![]()
Protein Composition
Amino Acids
Amino acids are the monomers that comprise proteins (Fig 21.3). Each amino acid has the same fundamental structure, which consists of a central carbon atom, or the alpha (α) carbon, bonded to an amino group (NH2), a carboxyl group (COOH), and to a hydrogen atom. Every amino acid also has another atom or group of atoms bonded to the central atom known as the R group (Figure 21.3).
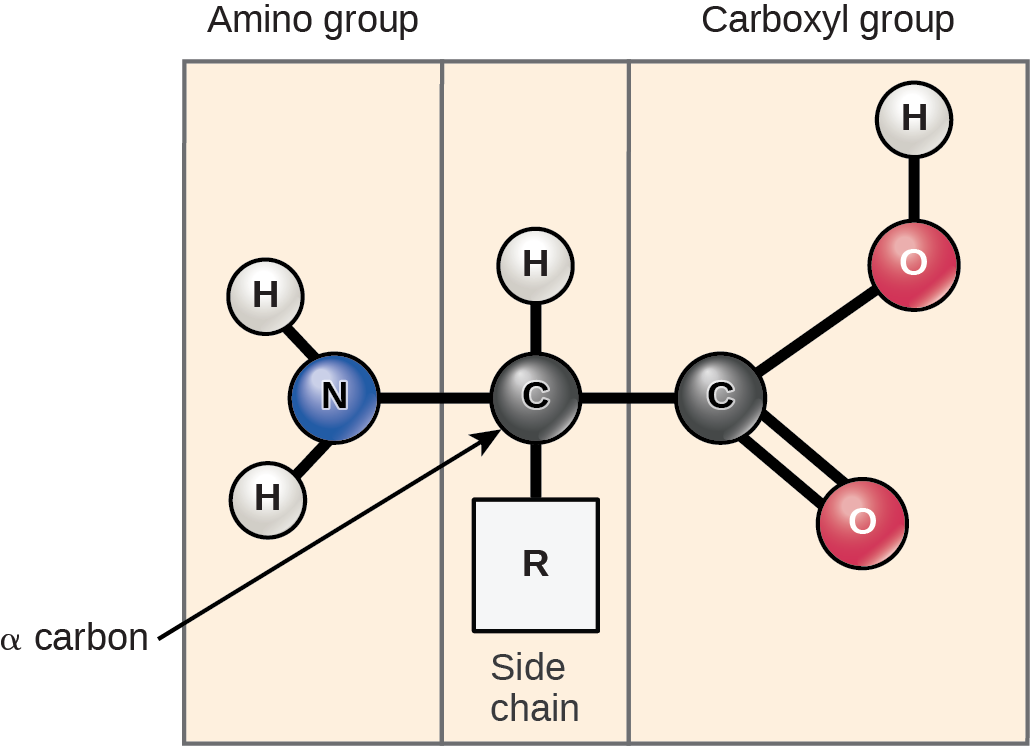
Scientists use the name “amino acid” because these acids contain both amino group and carboxyl-acid-group in their basic structure. As we mentioned, there are 20 common amino acids present in proteins. Nine of these are essential amino acids in humans because the human body cannot produce them and we obtain them from our diet. For each amino acid, the R group (or side chain) is different (Figure 21.4). Proline is an exception to the amino acid’s standard structure since its R group actually connects to the amino group, forming a ringlike structure (Figure 21.4).
Reading question #1
The component of the amino acid that makes each one unique is called:
A. the R group (or side group)
B. the carboxyl group
C. the central carbon
D. the amino group
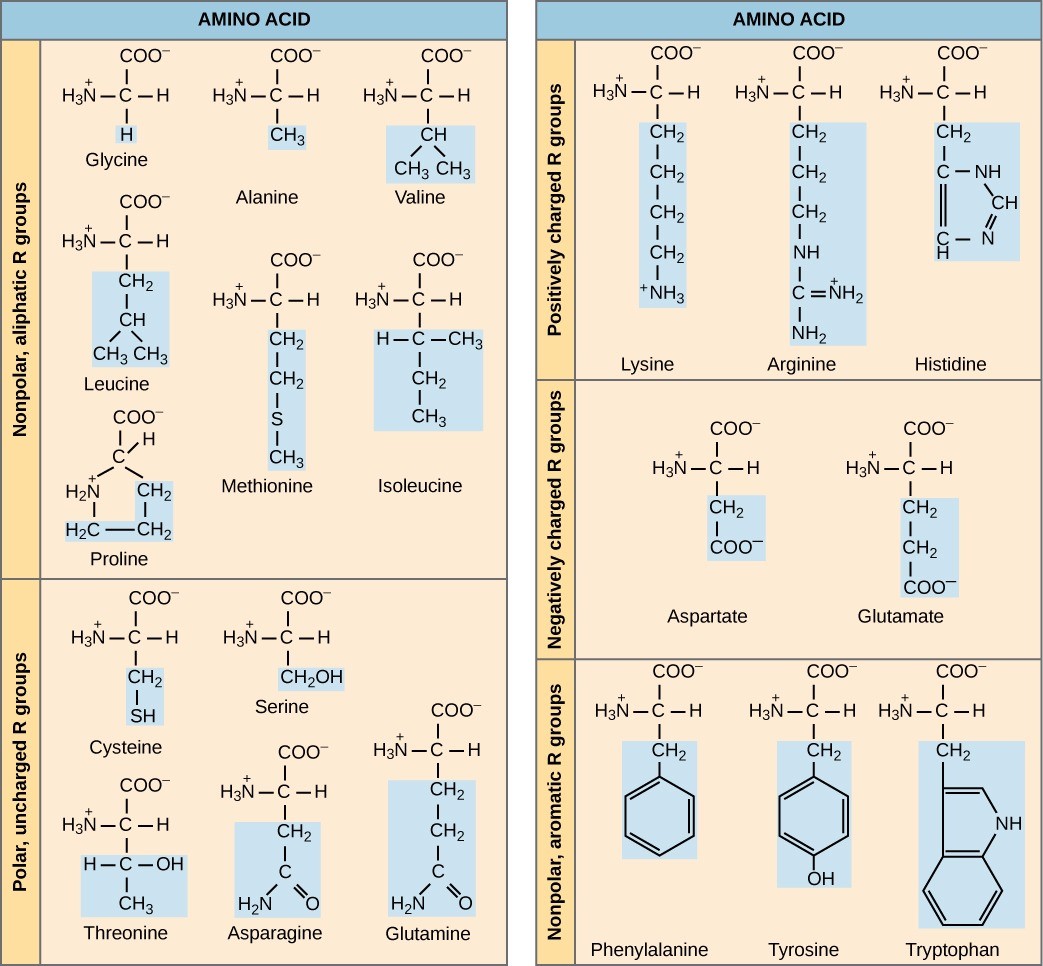
The chemical nature of the side chain determines the amino acid’s nature (that is, whether it is acidic, basic, polar, or nonpolar). For example, amino acids such as valine, methionine, and alanine are nonpolar and thus hydrophobic in nature, while amino acids such as serine, threonine, and cysteine are polar and thus have hydrophilic side chains. The side chains of lysine and arginine are positively charged, and therefore these amino acids are also basic amino acids.
A single upper case letter or a three-letter abbreviation represents amino acids. For example, the letter V or the three-letter symbol val represent valine. Just as some fatty acids are essential to a diet, some amino acids also are necessary. These essential amino acids in humans include isoleucine, leucine, and cysteine. Essential amino acids refer to those necessary to build proteins in the body, but not those that the body produces. Which amino acids are essential varies from organism to organism.
The sequence and the number of amino acids ultimately determine the protein’s shape, size, and function. A peptide bond (a type of covalent bond) attaches each amino acid to the next one in the chain. One amino acid’s carboxyl group and the incoming amino acid’s amino group combine, releasing a water molecule. The resulting bond is the peptide bond (Figure 21.5).
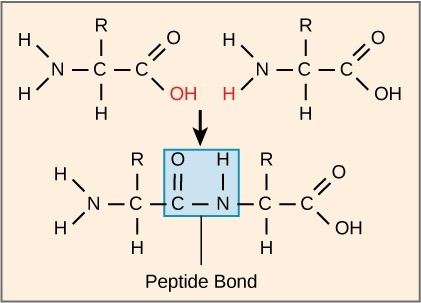
The products that such linkages form are peptides. As more amino acids join to this growing chain, the resulting chain is a polypeptide. Each polypeptide has a free amino group at one end. This end the N terminal, or the amino terminal, and the other end has a free carboxyl group, called the C or carboxyl terminal. While the terms polypeptide and protein are sometimes used interchangeably, a polypeptide is technically a polymer of amino acids, whereas the term protein is used for a polypeptide or polypeptides that have combined together, often have bound non-peptide prosthetic groups, have a distinct shape, and have a unique function. After protein synthesis (translation), most proteins are modified. These are known as post-translational modifications. They may undergo cleavage, phosphorylation, or may require adding other chemical groups. Only after these modifications is the protein completely functional.
Reading Question #2
What determines the nature of an amino acid, whether it is acidic, basic, polar, or nonpolar?
A. The number of amino acids in the polypeptide chain
B. The sequence of amino acids in the polypeptide chain
C. The size of the polypeptide chain
D. The chemical nature of its side chain (R group)
Reading Question #3
What is the name of the end of a polypeptide chain that has a free carboxyl group?
A. N terminal
B. Amino terminal
C. C terminal
D. Carbon terminal
Protein Structure
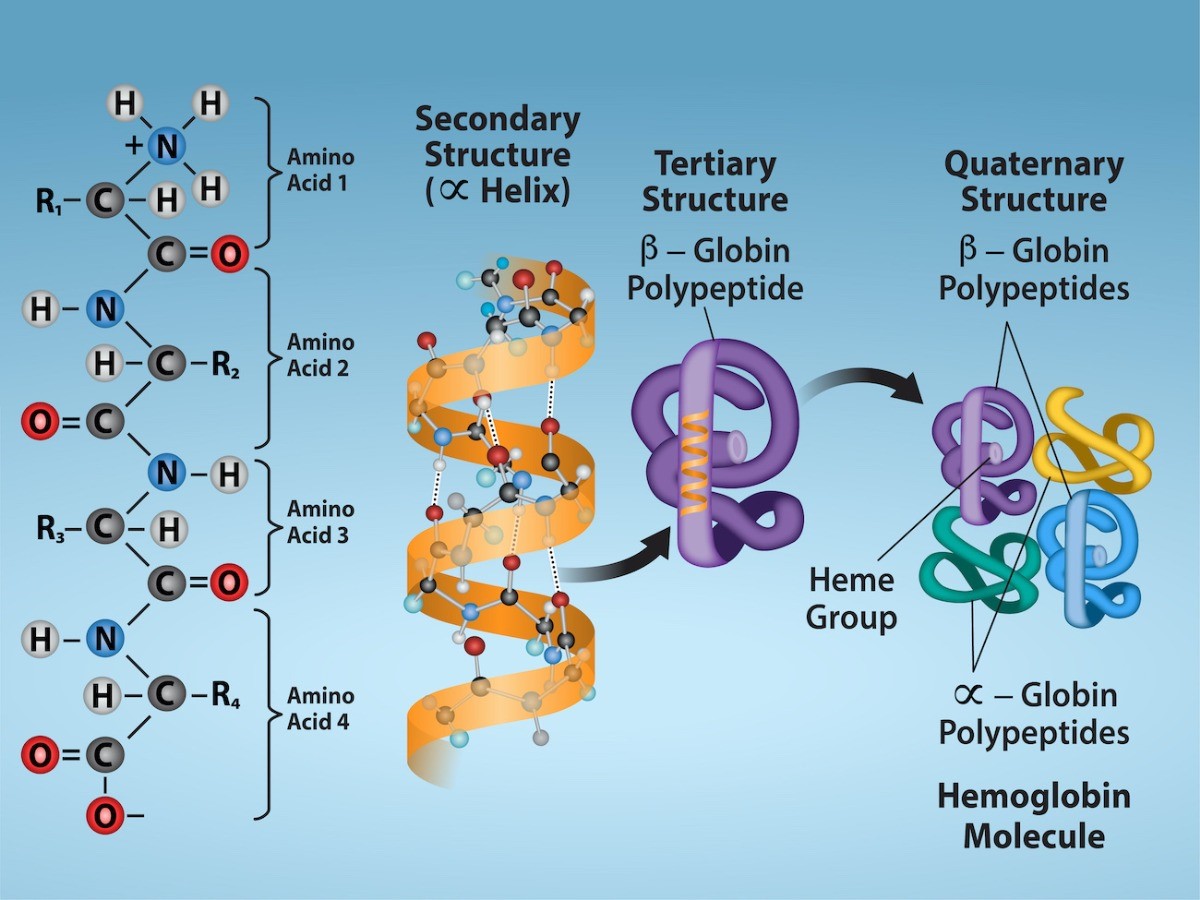
A protein’s shape is critical to its function. For example, an enzyme can bind to a specific substrate at an active site. If this active site is altered because of local changes or changes in overall protein structure, the enzyme may be unable to bind to the substrate. To understand how the protein gets its final shape or conformation, we need to understand the four levels of protein structure: primary, secondary, tertiary, and quaternary.
Primary Structure
Amino acids’ unique sequence in a polypeptide chain is its primary structure. For example, the pancreatic hormone insulin has two polypeptide chains, A and B, and they are linked together by disulfide bonds. The N terminal amino acid of the A chain is glycine; whereas, the C terminal amino acid is asparagine (Figure 21.6). The amino acid sequences in the A and B chains are unique to insulin.
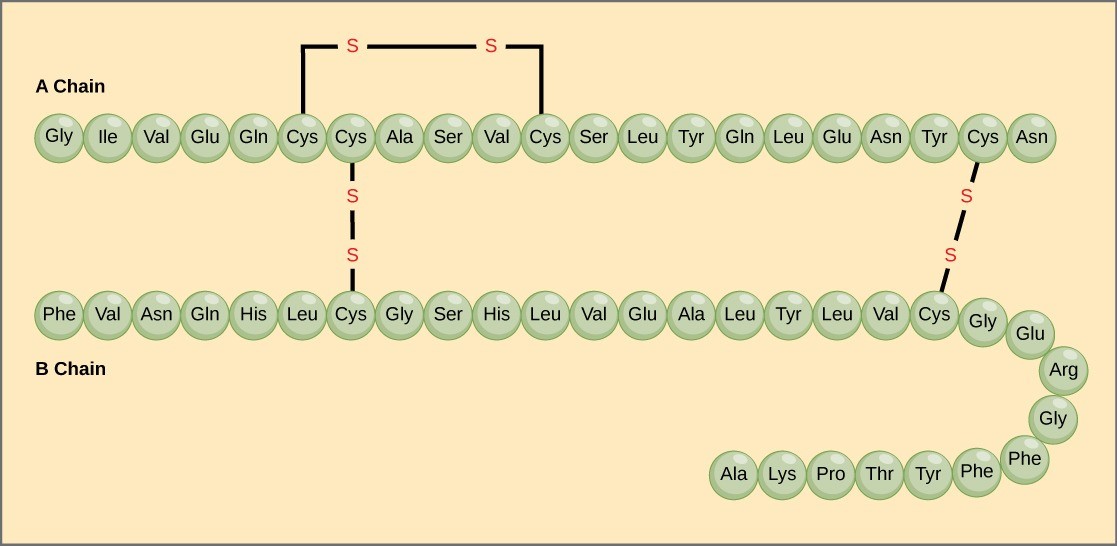
The gene encoding the protein ultimately determines the unique sequence for every protein. A change in nucleotide sequence of the gene’s coding region may lead to adding a different amino acid to the growing polypeptide chain, causing a change in protein structure and function. In sickle cell anemia, the hemoglobin β chain (a small portion of which is shown in Figure 21.7) has a single amino acid substitution, causing a change in protein structure and function. Specifically, valine in the β chain substitutes the amino acid glutamate. What is most remarkable to consider is that a hemoglobin molecule is comprised of two alpha and two beta chains that each consist of about 150 amino acids. The molecule, therefore, has about 600 amino acids. The structural difference between a normal hemoglobin molecule and a sickle cell molecule—which dramatically decreases life expectancy—is a single amino acid of the 600. What is even more remarkable is that three nucleotides each encode those 600 amino acids, and a single base change (point mutation), 1 in 1800 bases causes the mutation.
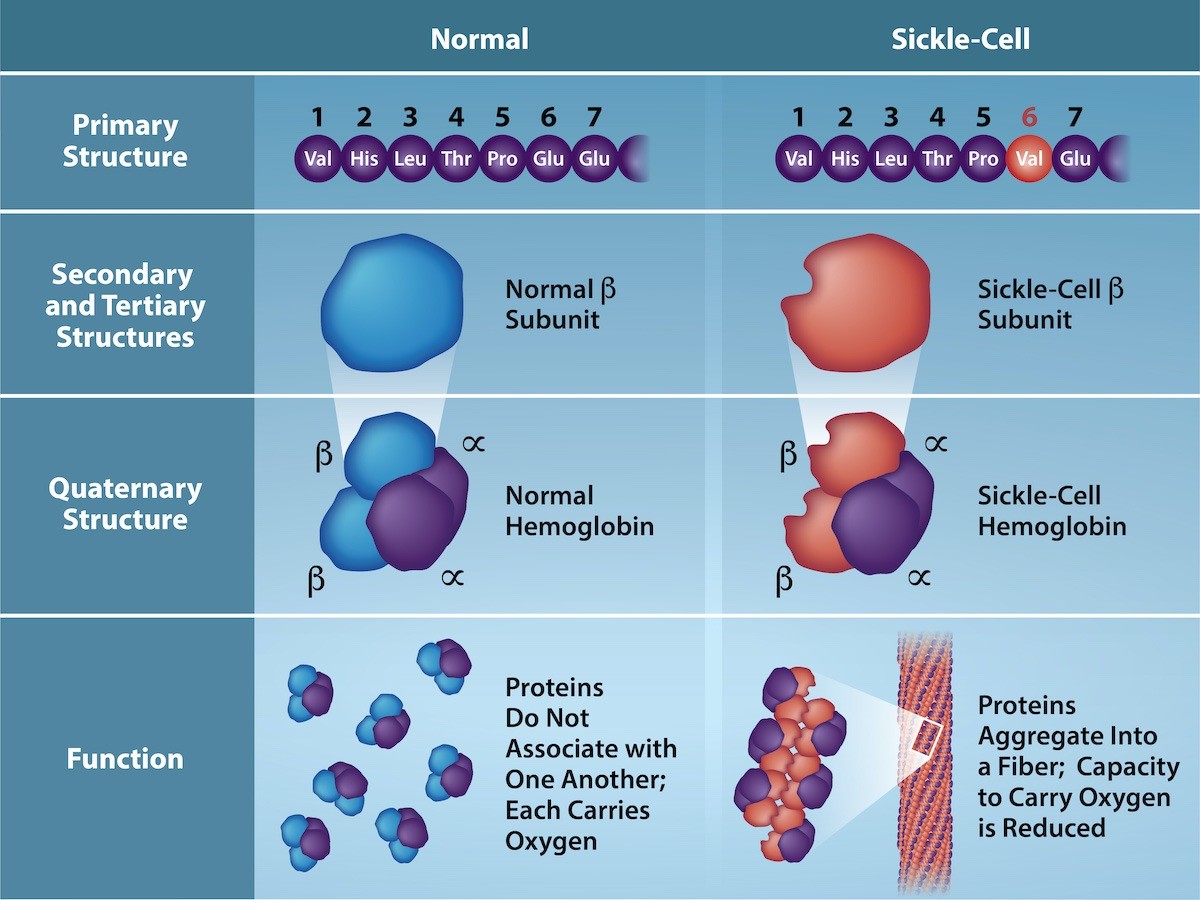
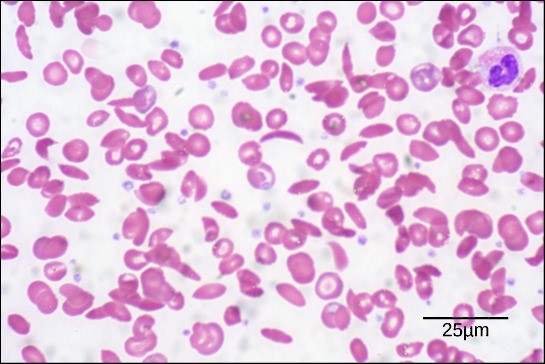
Because of this change of one amino acid in the chain, hemoglobin molecules form long fibers that distort the usual disc-shape of the red blood cells and causes them to assume a crescent or “sickle” shape, which clogs blood vessels (Figure 21.8). This can lead to myriad serious health problems such as breathlessness, dizziness, headaches, and abdominal pain for those affected by this disease. William Warrick Cardozo showed that sickle-cell anemia is an inherited disorder, meaning that the difference in the specific gene’s encoding region is passed down from parents to children. As you will learn in the genetics unit, the inheritance of such traits is determined by a combination of genes from both parents, and these very small differences can have significant impacts on organisms.
Secondary Structure
The local folding of the polypeptide in some regions gives rise to the secondary structure of the protein. The most common are the α-helix and β-pleated sheet structures (Figure 21.9). Both structures are held in shape by hydrogen bonds. The hydrogen bonds form between the oxygen atom in the carbonyl group in one amino acid and another amino acid that is four amino acids farther along the chain.
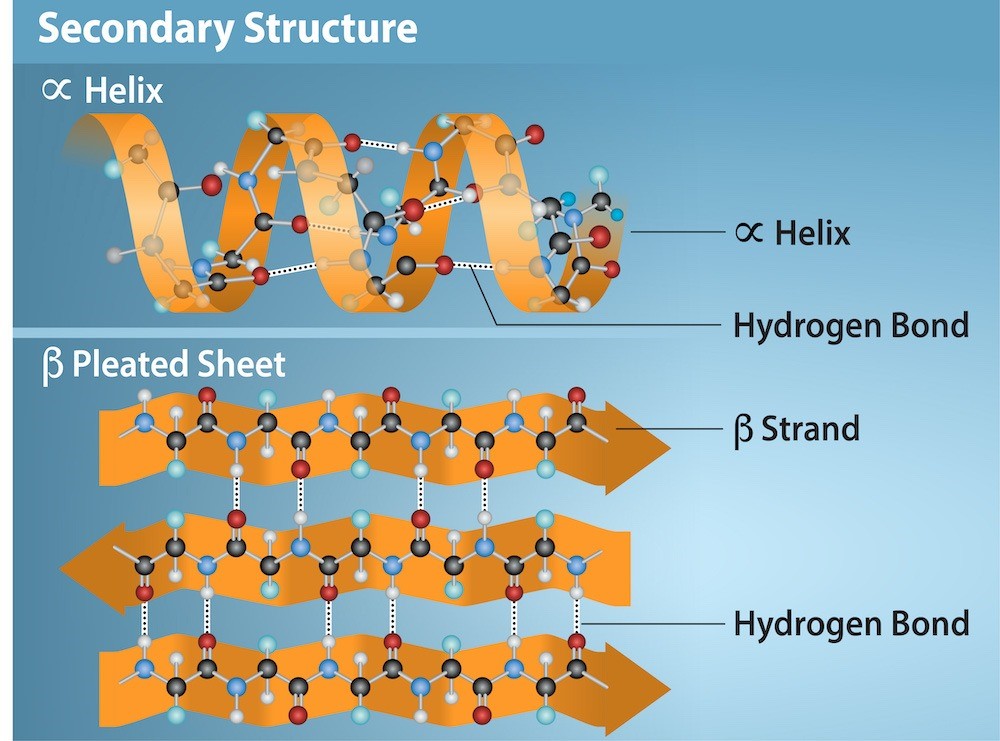
The polypeptide’s R groups (the variant groups) protrude out from the α-helix chain. In the β-pleated sheet, hydrogen bonding between atoms on the polypeptide chain’s backbone form the “pleats.” The R groups are attached to the carbons and extend above and below the pleat’s folds. The pleated segments align parallel or antiparallel to each other, and hydrogen bonds form between the partially positive hydrogen atom in the amino group and the partially negative oxygen atom in the peptide backbone’s carbonyl group. The α-helix and β-pleated sheet structures are in most globular and fibrous proteins and they play an important structural role.
Tertiary Structure
The polypeptide’s unique three-dimensional structure is its tertiary structure (Figure 21.10). This structure is in part due to chemical interactions at work on the polypeptide chain. Primarily, the interactions among R groups create the protein’s complex three-dimensional tertiary structure. The nature of the R groups in the amino acids involved can counteract forming the hydrogen bonds we described for standard secondary structures. For example, R groups with the same charges repel each other and those with opposite charges are attracted to each other (and form ionic bonds). When protein folding takes place, the non-polar amino acids’ hydrophobic R groups lie in the protein’s interior; whereas, the hydrophilic R groups lie on the outside. Scientists also call the former interaction types hydrophobic interactions. Interaction between cysteine side chains forms disulfide linkages in the presence of oxygen, the only covalent bond that forms during protein folding.
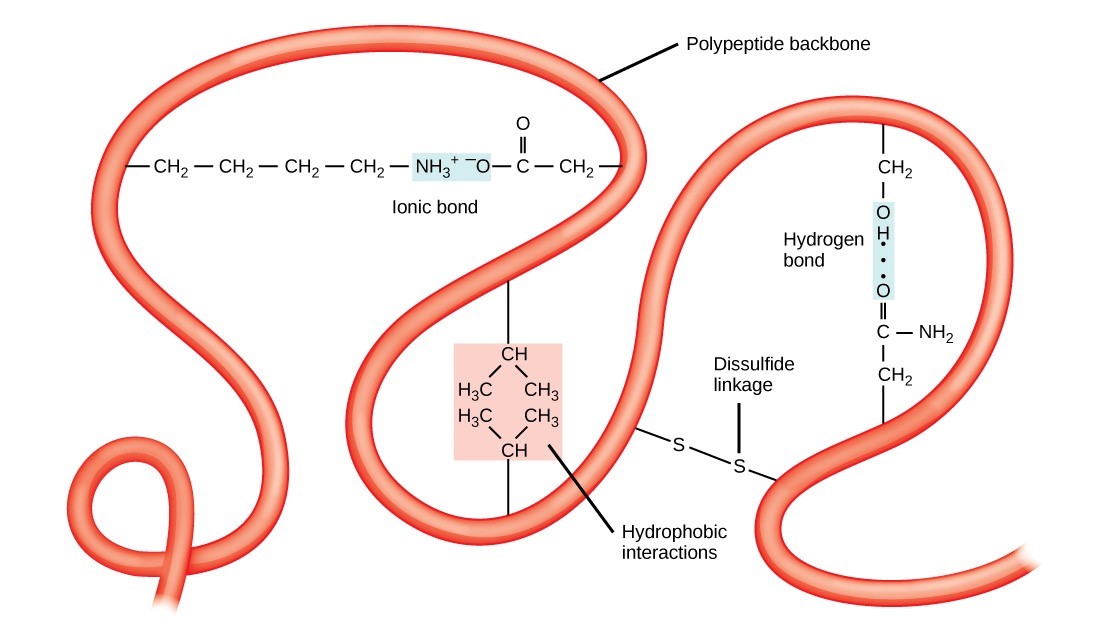
All of these interactions, weak and strong, determine the protein’s final three-dimensional shape. When a protein loses its three-dimensional shape, it may no longer be functional.
Quaternary Structure
Some proteins form from several polypeptides, called subunits, and the interaction of these subunits forms the quaternary structure. Weak interactions between the subunits help to stabilize the overall structure. For example, insulin (a globular protein) has a combination of hydrogen and disulfide bonds that cause it to mostly clump into a ball shape. Insulin starts out as a single polypeptide and loses some internal sequences in the presence of post-translational modification after forming the disulfide linkages that hold the remaining chains together. Silk (a fibrous protein), however, has a β-pleated sheet structure that is the result of hydrogen bonding between different chains.
Figure 22.11 illustrates the four levels of protein structure: primary, secondary, tertiary, and quaternary.
Research Connection: Marie Maynard Daly
Dr. Marie Maynard Daly was the first African-American woman to earn a doctorate degree in chemistry (Fig 21.12). Throughout her career, she made significant contributions to the scientific community’s understanding of proteins, amino acids, and other macromolecular building blocks (Jackson, 2022).

Most of her research focused on four topics: (1) the chemistry of histones, (2) protein synthesis, (3) the biochemistry of cholesterol and how it relates to hypertension and atherosclerosis, and (4) the uptake of creatine by muscle cells (Seifter, 2005). The studies she published focusing on protein synthesis were described as influential when the Nobel prize was awarded for the discovery of the structure of DNA. In addition to her research contributions, Dr. Daly encouraged minority students to enter the physical scientists by establishing a scholarship program at Queens College (Jackson, 2022).
Reading Question #4
Which level of protein structure is formed by hydrogen bonding between atoms in the backbone of a polypeptide chain?
A. Primary
B. Secondary
C. Tertiary
D. Quaternary
Denaturation and Protein Folding
Each protein has its own unique sequence and shape that chemical interactions hold together. If the protein is subjected to changes in temperature, pH, or exposure to chemicals, the protein structure may change, losing its shape without losing its primary sequence – this is called denaturation. Denaturation is often reversible because the polypeptide’s primary structure is conserved in the process. If the denaturing agent (i.e., high temperature or changed pH) is removed, the protein can often resume its function. Sometimes denaturation is irreversible, leading to loss of function. One example of irreversible protein denaturation is frying an egg. The albumin protein in the liquid egg white denatures when exposed to high temperatures. Not all proteins denature at high temperatures. For instance, bacteria that survive in hot springs have proteins that function at temperatures close to boiling. The stomach is also very acidic (ie., has a low pH) and denatures proteins as part of the digestion process; however, the stomach’s digestive enzymes retain their activity under these conditions.
Protein folding is critical to its function. Scientists originally thought that the proteins themselves were responsible for the folding process. Only recently researchers discovered that often they receive assistance in the folding process from protein helpers, or chaperones (or chaperonins) that associate with the target protein during the folding process. They act by preventing polypeptide aggregation that comprise the complete protein structure, and they disassociate from the protein once the target protein is folded.
Reading Question #5
What is the role of protein chaperones in the folding process of a protein?
A. They form hydrogen bonds with the protein to stabilize its structure.
B. They prevent polypeptide aggregation during folding.
C. They break disulfide linkages to facilitate protein folding.
D. They help in forming the secondary structure of the protein.
References and Acknowledgements
Jackson, N. (2022, February 25). Understanding of the macromolecular building blocks, with Dr. Marie Maynard Daly. Harvard School of Public Health. Retrieved from https://www.hsph.harvard.edu/molecular-
metabolism/2022/02/25/understanding-of-the-macromolecular-building-blocks-with-dr-marie-maynard-daly/
Seifter, S. (2005). Marie M. Daly PhD Memorial Celebration. Einstein Magazine Winter. Retrieved from https://www.einsteinmed.edu/education/phd/current-students/events/Marie-Daly-Celebration.aspx
Text adapted from: Clark, M.A., Douglas, M., and Choi, J. (2018, March 28). Biology 2e. OpenStax. Retrieved from https://openstax.org/books/biology-2e/pages/1-introduction
Adapted from Boundless. (2023). General Biology. LibreTexts. Retrieved from https://bio.libretexts.org/Bookshelves/Introductory_and_General_Biology/Book%3A_General_Biology_(Boundless).
Adapted from Choi, J., Clark, M. A., & Douglas, M. (2022) Biology 2e Part I, 2nd edition. by LOUIS: The Louisiana Library Network. Retrieved from https://louis.pressbooks.pub/generalbiology1leclab/

