14 Chapter 14: Transcription and RNA Processing
Lisa Limeri; Joshua Reid; and rocksher
Learning Objectives
By the end of this section, you will be able to do the following:
-
Explain the purpose of each modification made during RNA processing and predict the consequences of each modification failing.

Transcription: DNA Encodes for RNA
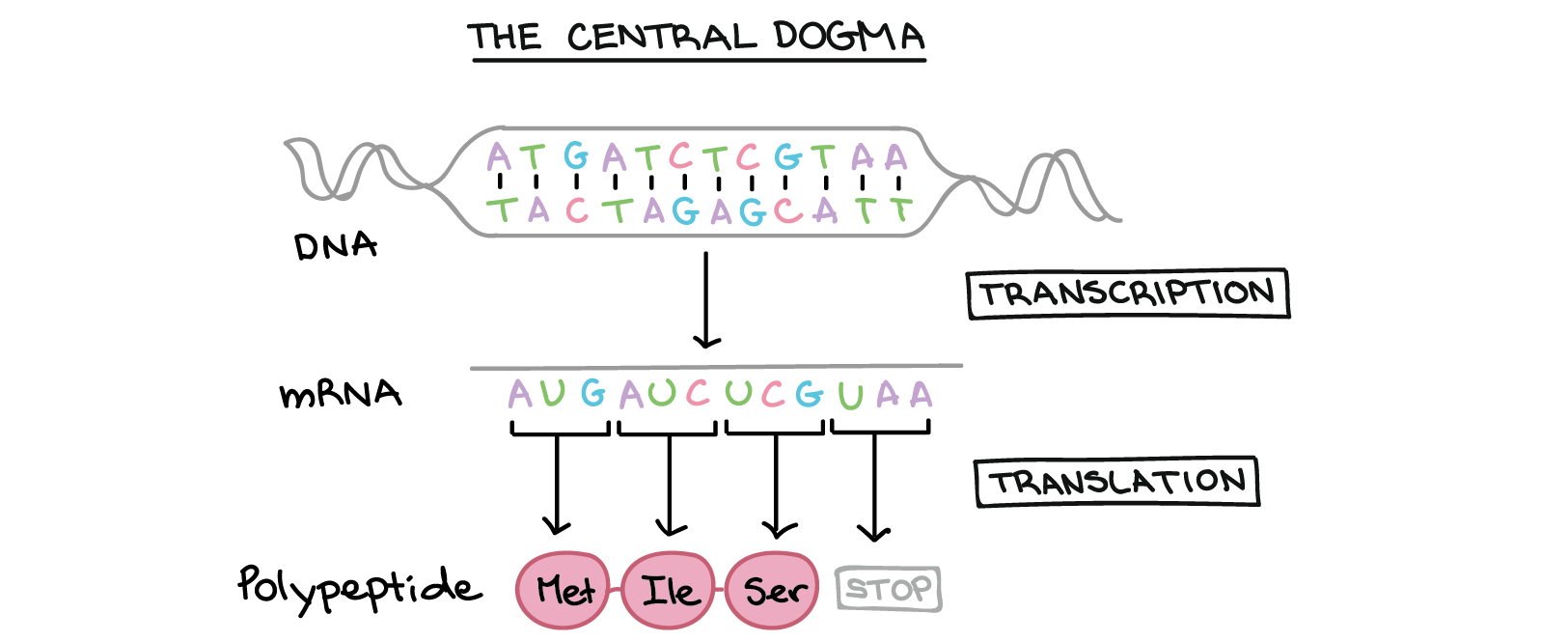
The flow of genetic information in cells from DNA to mRNA is accomplished by a process known as transcription (Fig 16.1). This first step in the central dogma of biology serves a few purposes. Because the information stored in DNA is so central to cellular function, it makes sense that the cell would make temporary copies of the information, in the form of mRNA, while keeping the DNA itself intact and protected in the nucleus of the cell. Additionally, the production of mRNA also amplifies the information; one gene gives rise to thousands of copies of mRNA templates, each of which may be used to produce large amounts of protein quickly through the actions of ribosomes.
Copying DNA to RNA is relatively straightforward, with one nucleotide being added to the mRNA strand for every nucleotide read in the DNA strand using the base-pairing rules (Table 16.1 and Figure 16.2). The resulting mRNA contains the genetic information for the gene that was copied. Eukaryotic pre-mRNAs undergo extensive processing after transcription but before translation.
|
Base found in DNA |
Base inserted into RNA |
|
A |
U |
|
T |
A |
|
C |
G |
|
G |
C |

Reading Question #1
What is the purpose of transcription in the central dogma of biology?
A. To synthesize proteins directly from DNA
B. To convert RNA into DNA
C. To create temporary copies of genetic information in the form of mRNA
D. To produce ribosomes for protein synthesis
Eukaryotic Transcription
Prokaryotes and eukaryotes perform fundamentally the same process of transcription, with a few key differences. The most important difference between prokaryote and eukaryote transcription is due to the membrane-bound nucleus and organelles found within the eukaryotic cell. With the genes bound in a nucleus, the eukaryotic cell must be able to transport its mRNA to the cytoplasm and must protect its mRNA from degrading before it is translated. Eukaryotes also employ three different transcription enzymes called RNA polymerases that each transcribe a different subset of genes.
The process of transcription is divided into three distinct stages: initiation, elongation, and termination.
Transcription Initiation
Transcription initiation requires the binding of RNA polymerase to a region of DNA called the promoter. Initiation starts when the double-stranded DNA template is separated, through the breaking of hydrogen bonds, to form the transcription bubble. This step commits the RNA polymerase to move down the DNA template and add nucleotides to the growing RNA chain. Eukaryotes require three distinct RNA polymerases, as well as several proteins called transcription factors, to initiate transcription at the promoter. Each eukaryotic polymerase requires a distinct set of transcription factors to bring it to the DNA template.
RNA polymerase II is responsible for transcribing the overwhelming majority of eukaryotic genes (Table 16.2). This enzyme is located in the nucleus and synthesizes all protein-coding nuclear pre-mRNAs.
RNA polymerase I is located in the nucleolus, a specialized nuclear substructure in which ribosomal RNA (rRNA) is transcribed, processed, and assembled into ribosomes (Table 16.2). The rRNA molecules are considered structural RNAs because they have a cellular role but are not translated into protein. The rRNAs are components of the ribosome and are essential to the process of translation.
RNA polymerase III is also located in the nucleus. This polymerase transcribes a variety of structural RNAs that includes the 5S pre-rRNA, transfer pre-RNAs (pre-tRNAs), and small nuclear pre-RNAs (Table 16.2). The tRNAs have a critical role in translation; they serve as the “adaptor molecules” between the mRNA template and the growing polypeptide chain. Small nuclear RNAs have a variety of functions, including splicing pre-mRNAs and regulating transcription factors.
|
RNA Polymerase |
Cellular Compartment |
Product of Transcription |
|
I |
Nucleolus |
All rRNAs except 5S rRNA |
|
II |
Nucleus |
All protein-coding nuclear pre- mRNAs |
|
III |
Nucleus |
5S rRNA, tRNAs, and small nuclear RNAs |
For the purposes of our discussion, we will focus on RNA polymerase II since it is responsible for the synthesis of mRNA used for protein synthesis.
Eukaryotic promoters contain a sequence of DNA is called the TATA box (TATAAA) on the coding strand of DNA. It is located at -25 to -35 bases relative to the initiation (+1) start site (Figure 16.3). The relative stability of A–T bonds is low (due to the two hydrogen bonds between A and T compared to three hydrogen bonds between G-C), making it easier for RNA Polymerase to locally unwind the double stranded DNA template in preparation for transcription.
To initiate transcription, eukaryotes assemble a complex of transcription factors required to recruit RNA polymerase II to a protein coding gene. Transcription factors that bind to the promoter are called basal transcription factors. These basal factors are all called TFII (for Transcription Factor/polymerase II) plus an additional letter (A-J). The core complex is TFIID, which includes a TATA-binding protein (TBP; Figure 16.3). The other transcription factors systematically fall into place on the DNA template, with each one further stabilizing the pre-initiation complex and contributing to the recruitment of RNA polymerase II.
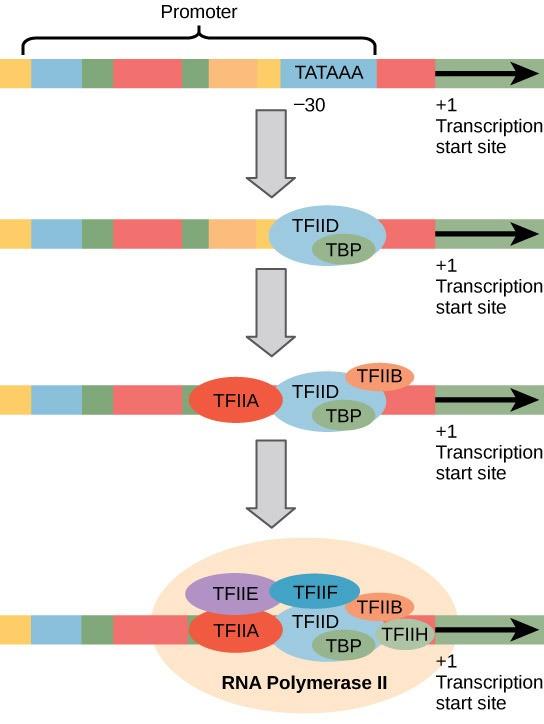
Some eukaryotic promoters also have a conserved CAAT box (GGCCAATCT) at approximately -80 upstream of the +1 site. Further upstream of the TATA box, eukaryotic promoters may also contain one or more GC-rich boxes (GGCG) or octamer boxes (ATTTGCAT). These elements bind cellular factors that increase the efficiency of transcription initiation and are often identified in more “active” genes that are constantly being expressed by the cell.
Basal transcription factors are crucial in the formation of a pre-initiation complex on the DNA template that subsequently recruits RNA polymerase II for transcription initiation. The complexity of eukaryotic transcription does not end with the polymerases and promoters. An army of other transcription factors, which bind to upstream DNA sequences known as enhancers and silencers, also help to regulate the frequency with which pre-mRNA is synthesized from a gene. Enhancers and silencers affect the efficiency of transcription but are not necessary for transcription to proceed.
Research Connection: Transcription Factors and “Laziness”
Most of the time, DNA replication occurs predictably, with nucleotides pairing together their complementary base pair (A with T, and C with G). However, on occasion, polymerase will pair a nucleotide with the wrong partner, resulting in a mismatched section of DNA. A team of researchers led by Dr. Raluca Gordân found that transcription factor proteins had stronger bonds with sections of DNA containing mismatched nucleotide pairs. After examining their experimental results, the researchers concluded that this strong bond is largely the result of protein “laziness”! “When a transcription factor protein binds to DNA, it must spend energy distorting the site, for example by bending the DNA to its will. However, mismatched sections of DNA are already distorted, so the transcription factor protein has to do less work” (Duke University, 2020). In her future research projects, Dr. Gordân hopes to investigate how this phenomenon relates to disease development. This strong bond between proteins and mismatched sections of DNA could interfere with the repair of nucleotide mismatches, leading to accumulated mutations that could progress into diseases such as cancer or neurodegeneration (Afek et al., 2020).
Reading Question #2
Which enzyme is responsible for transcribing the majority of eukaryotic genes, producing all protein-coding nuclear pre-mRNAs?
A. RNA polymerase I
B. RNA polymerase II
C. RNA polymerase III
D. RNA polymerase IV
Transcription Elongation
Following the formation of the pre-initiation complex, the polymerase is released from the other transcription factors, and elongation proceeds, with the polymerase synthesizing pre-mRNA in the 5’ to 3’ direction (Fig 16.4). As discussed previously, RNA polymerase II transcribes the major share of eukaryotic genes, so in this section we will focus on how this polymerase accomplishes elongation and termination.

The DNA template used in eukaryotic transcription is complex due to the fact that when eukaryotic cells are not dividing, their genes exist as a diffuse mass of DNA and proteins called chromatin. The DNA is tightly packaged around charged histone proteins at repeated intervals. These DNA–histone complexes, collectively called nucleosomes, are regularly spaced and include 146 nucleotides of DNA wound around eight histones like thread around a spool. For RNA synthesis to occur, the transcription machinery needs to move histones out of the way every time it encounters a nucleosome. This is accomplished by a special protein complex called FACT, which stands for “facilitates chromatin transcription.” This complex pulls histones away from the DNA template as the polymerase moves along it. Once the pre-mRNA is synthesized, the FACT complex replaces the histones to recreate the nucleosomes.
Transcription Termination
The termination of transcription is different for the different polymerases. Elongation by RNA polymerase II in eukaryotes takes place 1,000 to 2,000 nucleotides beyond the end of the gene being transcribed. This premRNA tail is subsequently removed by cleavage during mRNA processing. On the other hand, RNA polymerases I and III require termination signals. Genes transcribed by RNA polymerase I contain a specific 18-nucleotide sequence that is recognized by a termination protein. The process of termination in RNA polymerase III involves the formation of a double-stranded structure at the end of the RNA known as an RNA hairpin. This hairpin causes the disassociation of RNA polymerase with the DNA template and the release of primary transcript.
Reading Question #3
Which polymerase requires a specific termination sequence in the DNA to signal the end of transcription?
A. RNA polymerase I
B. RNA polymerase II
C. RNA polymerase III
D. RNA polymerase IV

RNA Processing in Eukaryotes
After transcription, eukaryotic pre-mRNAs must undergo several processing steps to become mature mRNA before they can be translated. The additional steps involved in eukaryotic mRNA maturation create a molecule with a longer half-life; eukaryotic mRNAs last for several hours. Eukaryotic (and prokaryotic) tRNAs and rRNAs also undergo processing before they can function as components in the protein-synthesis machinery.
Pre-mRNAs are first coated in RNA-stabilizing proteins; these protect the pre-mRNA from degradation while it is processed and exported out of the nucleus. The three most important steps of pre-mRNA processing are the addition of stabilizing and signaling factors at the 5’ and 3’ ends of the molecule, and the removal of the non-coding dNA segments (called introns) through splicing. In rare cases, the mRNA transcript can be “edited” after it is transcribed (see the evolution connection below).
Splicing
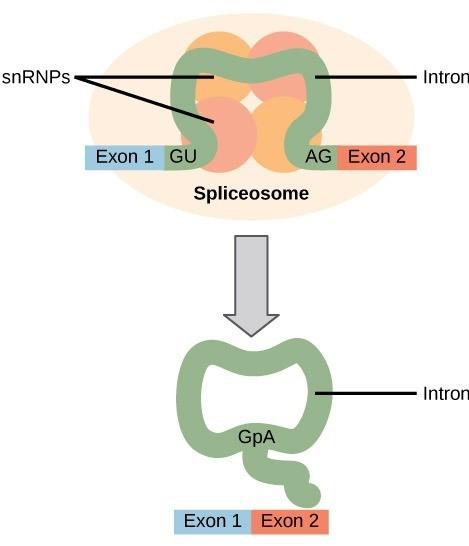
Eukaryotic genes are composed of exons, which correspond to protein-coding sequences (ex-on signifies that they are expressed), and intervening sequences called introns (int-ron denotes their intervening role), which may be involved in gene regulation but are removed from the pre-mRNA during processing. Intron sequences in mRNA do not encode functional proteins (16.5).
The discovery of introns came as a surprise to researchers in the 1970s who expected that pre-mRNAs would specify protein sequences without further processing, as they had observed in prokaryotes. The genes of higher eukaryotes very often contain one or more introns. These regions may correspond to regulatory sequences; however, the biological significance of having many introns or having very long introns in a gene is unclear. It is possible that introns slow down gene expression because it takes longer to transcribe pre-mRNAs with lots of introns. Alternatively, introns may be nonfunctional sequence remnants left over from the fusion of ancient genes throughout the course of evolution. This is supported by the fact that separate exons often encode separate protein subunits or domains. For the most part, the sequences of introns can be mutated without ultimately affecting the protein product. All of a pre-mRNA’s introns must be completely and precisely removed before protein synthesis. If the process errs by even a single nucleotide, the reading frame of the rejoined exons would shift, and the resulting protein would be dysfunctional. The process of removing introns and reconnecting exons is called splicing (Figure 16.6).
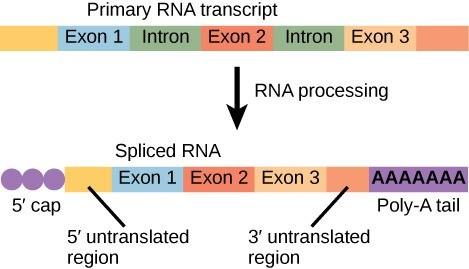
Introns are removed and degraded while the pre-mRNA is still in the nucleus. Splicing occurs by a sequence-specific mechanism that ensures introns will be removed and exons rejoined with the accuracy and precision of a single nucleotide. Although the intron itself is noncoding, the beginning and end of each intron is marked with specific nucleotides: GU at the 5’ end and AG at the 3’ end of the intron. The splicing of premRNAs is conducted by complexes of proteins and RNA molecules called spliceosomes (Fig 16.6). The splicing process is catalyzed by protein complexes called spliceosomes that are composed of proteins and RNA molecules called small nuclear RNAs (snRNAs). Spliceosomes recognize sequences at the 5’ and 3’ end of the intron. Note that more than 70 individual introns can be present, and each has to undergo the process of splicing—in addition to 5’ capping and the addition of a poly-A tail—just to generate a single, translatable mRNA molecule.
Addition of the 5′ G Cap
While the pre-mRNA is still being synthesized, a 7-methylguanosine cap is added to the 5’ end of the growing transcript by a phosphate linkage. This functional group protects the nascent mRNA from degradation. In addition, factors involved in protein synthesis recognize the cap to help initiate translation by ribosomes.
Addition of the 3′ Poly-A Tail
Once elongation is complete, the pre-mRNA is cleaved by an endonuclease between an AAUAAA consensus sequence and a GU-rich sequence, leaving the AAUAAA sequence on the pre-mRNA. An enzyme called poly-A polymerase then adds a string of approximately 200 A residues, called the poly-A tail. This modification further protects the pre-mRNA from degradation and is also the binding site for a protein necessary for exporting the processed mRNA to the cytoplasm.
Reading Question #4
What are the interrupted coding sequences in eukaryotic pre-mRNA called?
A. Exons
B. Introns
C. tRNAs
D. rRNAs
RNA Editing
The trypanosomes are a group of protozoa that include the pathogen Trypanosoma brucei, which causes nagana in cattle and sleeping sickness in humans throughout great areas of Africa. The trypanosome is carried by biting flies in the genus Glossina (commonly called tsetse flies). Trypanosomes, and virtually all other eukaryotes, have organelles called mitochondria that supply the cell with chemical energy. Mitochondria are organelles that express their own DNA and are believed to be the remnants of a symbiotic relationship between a eukaryote and an engulfed prokaryote. The mitochondrial DNA of trypanosomes exhibit an interesting exception to the central dogma: their pre-mRNAs do not have the correct information to specify a functional protein. Usually, this is because the mRNA is missing several U nucleotides. The cell performs an additional RNA processing step called RNA editing to remedy this.
Other genes in the mitochondrial genome encode 40- to 80-nucleotide guide RNAs. One or more of these molecules interacts by complementary base pairing with some of the nucleotides in the pre-mRNA transcript. However, the guide RNA has more A nucleotides than the pre-mRNA has U nucleotides with which to bind. In these regions, the guide RNA loops out. The 3’ ends of guide RNAs have a long poly-U tail, and these U bases are inserted in regions of the pre-mRNA transcript at which the guide RNAs are looped. This process is entirely mediated by RNA molecules. That is, guide RNAs—rather than proteins—serve as the catalysts in RNA editing.
RNA editing is not just a phenomenon of trypanosomes. In the mitochondria of some plants, almost all pre-mRNAs are edited. RNA editing has also been identified in mammals such as rats, rabbits, and even humans. What could be the evolutionary reason for this additional step in pre-mRNA processing? One possibility is that the mitochondria, being remnants of ancient prokaryotes, have an equally ancient RNA-based method for regulating gene expression. In support of this hypothesis, edits made to pre-mRNAs differ depending on cellular conditions. Although speculative, the process of RNA editing may be a holdover from a primordial time when RNA molecules, instead of proteins, were responsible for catalyzing reactions.
Reading Question #5
What is the role of guide RNAs in the RNA editing process of trypanosomes?
A. Guide RNAs serve as catalysts to remove introns during splicing.
B. Guide RNAs add stabilizing and signaling factors to the pre-mRNA.
C. Guide RNAs interact with pre-mRNA transcripts to insert U nucleotides where needed.
D. Guide RNAs protect the nascent mRNA from degradation during processing.
References and Acknowledgements
Afek, A., Shi, H., Rangadurai, A., Sahay, H., Senitzki, A. … Gordân, R. (2020). DNA mismatches reveal conformational penalties in protein–DNA recognition. Nature 587, 291–296. https://doi.org/10.1038/s41586020-2843-2
Duke University. (2020, October 21). Transcription factors may inadvertently lock in DNA mistakes: Binding to mismatched DNA takes less energy; may explain how regulatory mutations get locked in. ScienceDaily.
Retrieved from www.sciencedaily.com/releases/2020/10/201021112356.htm
adapted from Clark, M.A., Douglas, M., and Choi, J. (2018). Biology 2e. OpenStax. Retrieved from https://openstax.org/books/biology-2e/pages/1-introduction

